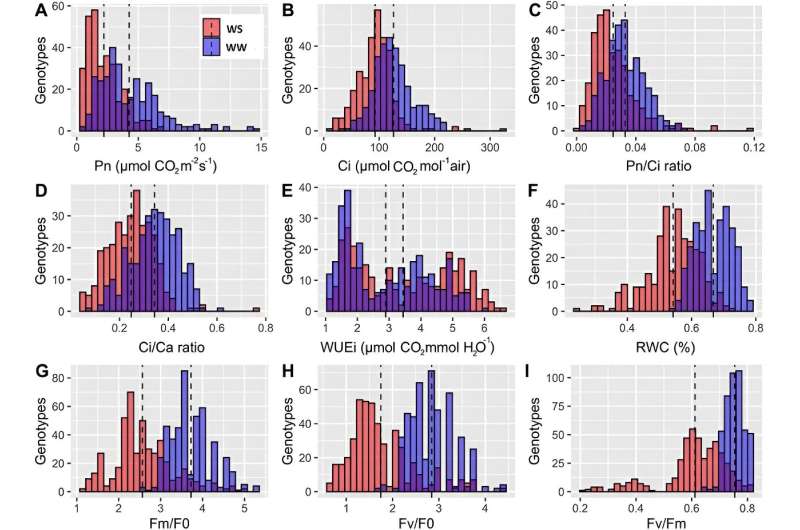
In the first year of the experiment, the distribution of photosynthetic-related traits of 6-month-old walnut plants under conditions of sufficient water (blue) and severe stress (red) and the overlap between them (purple). Credit: Horticulture Research
Walnut (Juglans regia L.) was domesticated in ancient Persia and is a globally cultivated nut crop. With global water scarcity, walnut production faces major challenges due to abiotic stresses, especially drought. Photosynthesis is a key physiological mechanism for adapting to abiotic stress and regulating plant development.
Research has mainly focused on the physiological effects of drought on walnuts, but its molecular mechanisms are not fully understood. Many new genomic tools have been widely used to study the genomic diversity and association mapping of walnut.
The Persian walnut reference genome and SNP genotyping array facilitate advanced genomic studies, including QTL mapping and genome-wide association studies (GWAS).
Technologies such as genotyping by sequencing (GBS) and restriction site-associated DNA sequencing (RADseq) have the potential to generate large marker datasets. However, many SNP markers with minor effects are always overlooked, and most of the genetic variants responsible for the trait remain hidden. Therefore, complementary approaches such as gene set enrichment analysis can provide a deeper understanding of the molecular mechanisms of walnut responses to drought stress.
June 2022, Horticulture Research published a research article titled “Genome-wide association analysis and pathway enrichment provide insights into the genetic basis of the photosynthetic response to drought stress in Persian walnut.”
First, this study explored natural variation in photosynthetic traits using 150 walnut families (1,500 seedlings) from major walnut-producing areas in Iran. They evaluated 30 photosynthesis-related traits, divided into two main categories (gas exchange and chlorophyll fluorescence measurements) under control, water stress and recovery conditions.
The results showed that the phenotypic variation among families was large, and most traits showed close to normal distribution. In addition to some gas exchange parameters (CI and CI/CA under drought recovery) and chlorophyll fluorescence measurements (VI Under drought conditions, FI and Fmedium size is recovering). Most photosynthetic traits showed substantial decreases under drought stress. Significant correlations between traits were observed under drought conditions.
To further explore key parameters and provide a comprehensive view of the relationships between traits within a population, principal component analysis (PCA) was performed on all photosynthetic traits of 140 families during stress. The first five components (PC15) cumulatively explained more than 90% of the total variation in photosynthetic traits across the entire panel under severe drought conditions, with the first component (PC1) significantly correlated with the moisture relationship parameters and F .V/Fmedium size index.
In SNP calling and population structure analyses, genomic data from 95 parent trees and 150 families were used. Array-scored SNPs were divided into six preset groups.
PCA and population structure estimates for each panel genotyped by GBS, and the panels were divided into four main clusters based on geographical location.
Genome-wide association studies (GWAS) using MArray, MGBS, and PGBS datasets identified 198 (34%), 228 (40%), and 152 (26%) genes associated with at least one photosynthesis-related trait across all conditions, respectively. Significant correlation. The BLASTX results then showed 578 and 1,543 significant and suggestive SNPs identified by GWAS.
Most SNPs were located within or near genes involved in the regulation of photosynthesis and drought tolerance. Gene set enrichment and network analysis extracted genes surrounding important and suggestive SNPs and identified multiple KEGG pathways and GO terms related to drought tolerance and photosynthesis. The enrichment pathways are related to metabolic processes such as carbohydrate metabolism, amino acid metabolism, lipid metabolism, energy metabolism and signal transduction.
Finally, protein-protein interactions identified central genes involved in photosynthesis and drought response, providing insights into the genetic control of photosynthetic traits under different conditions in Persian walnut.
Together, these findings highlight the potential of natural photosynthetic variation in walnut as a valuable resource for breeding and engineering more efficient photosynthesis, particularly in the context of climate change and water scarcity.
This study not only reveals the physiological and genetic adaptation of walnuts to drought, but also lays the foundation for future walnut breeding programs focused on improving drought tolerance.
More information:
Mohammad M Arab et al., Genome-wide association analysis and pathway enrichment provide insights into the genetic basis of the photosynthetic response to drought stress in Persian walnut, Horticulture Research (2022). DOI: 10.1093/hour/uhac124
Plant Phenomics provides
citation: Unlocking the genetic secrets of Persian walnut drought resistance (2023, December 21), Retrieved December 21, 2023, from https://phys.org/news/2023-12-genic-secrets-drought-resilience-persian. html
This document is protected by copyright. No part may be reproduced without written permission except in the interests of fair dealing for private study or research purposes. Content is for reference only.
#Genetic #secrets #Persian #walnuts #drought #resistance #revealed
Image Source : phys.org